Genetic determinants of the microbiome
Our lab is particularly interested in elucidating the role of host genetics in determining gut microbiome composition. Is the microbiome heritable? What host genetic variants are determine microbial abundance in the gut? How can we use host genetic variation to understand causal relationships between the microbiome and disease?

Microbiome GWAS in the Hutterites
photo: Kelly Hofer
Host genetics + microbiota interactions in the TwinsUK
Reviews and commentaries
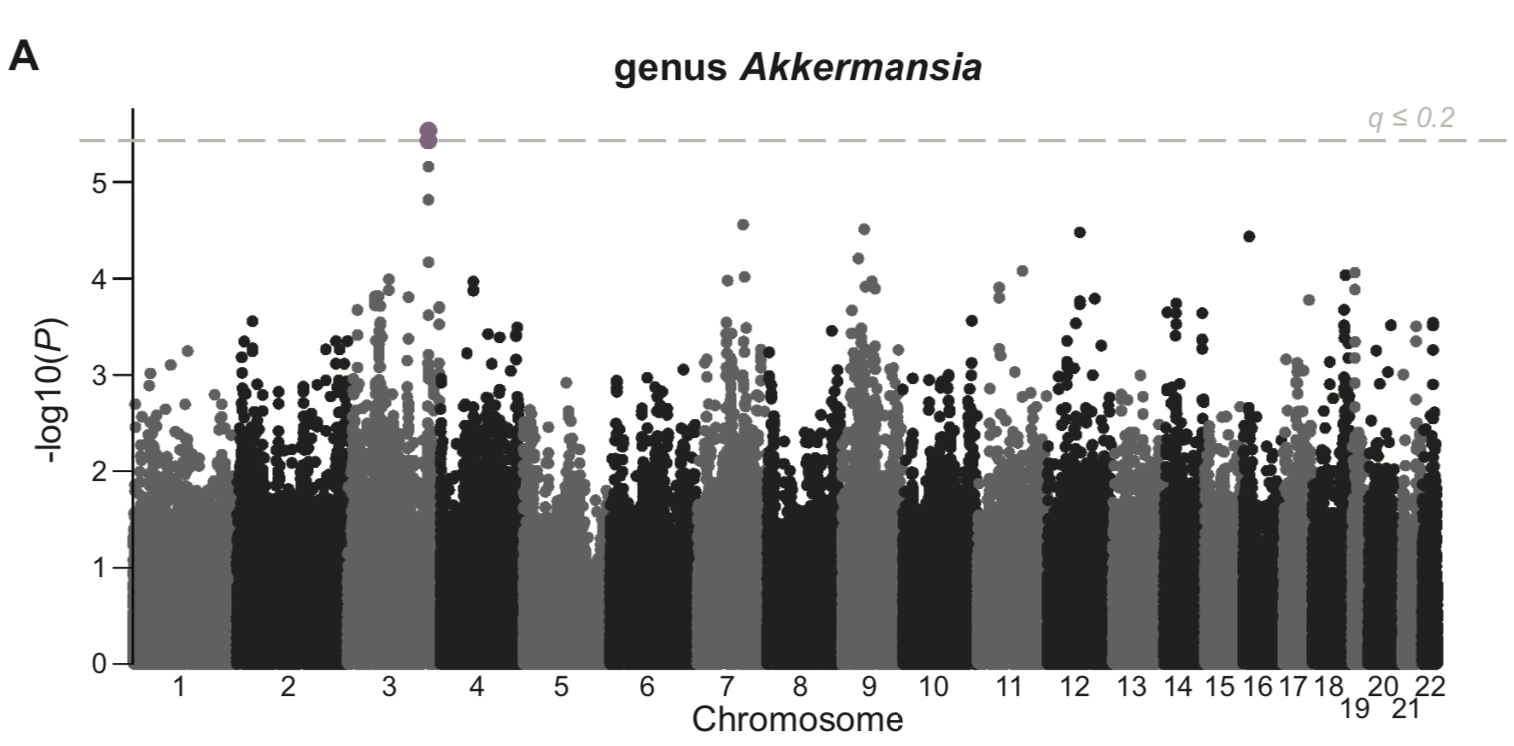
«Microbiome GWAS in the Hutterites»
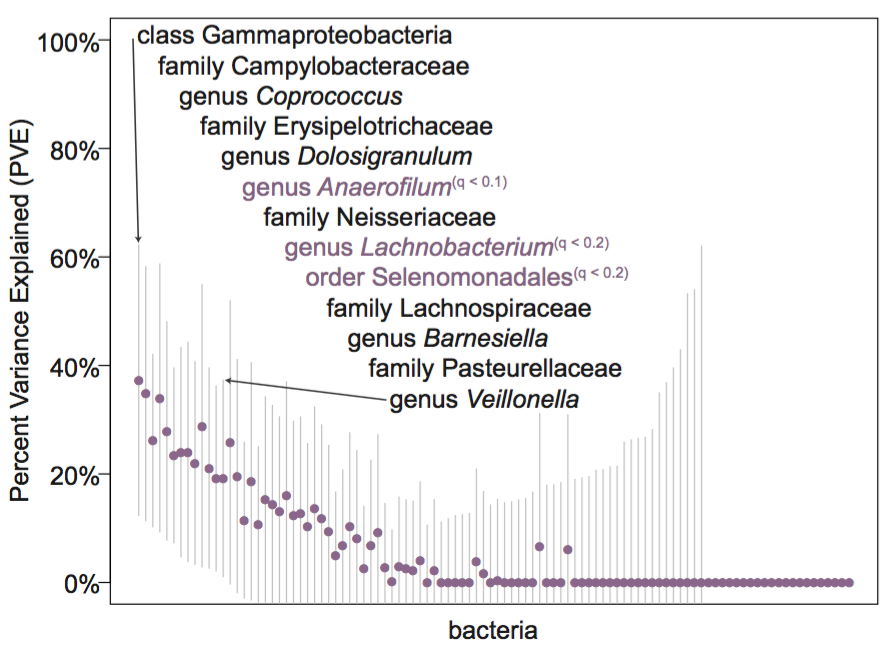
While many environmental factors are known to contribute to gut microbiome composition, the extent to which host genetic variation plays a role is unclear.
Using the same population as the seasonal study (the Hutterites), where inter-individual variation in diet is limited, I examined the role of sex and age as well as host genetics in determining the relative abundances of the most common bacteria in the gut.
To do so, I implemented methods that specifically account for the relatedness across individuals and examined genetic variation genome-wide.
While this is an important first step at examining host genetics, I really can’t state enough the limitations of the sample size in this study and the number of tests performed.
That being said, I do find bacteria that show evidence of heritability in my samples and have genome-wide (but not study-wide) significant associations with SNPs in the genome.
To read more, check out:
Genome-wide association studies of the human gut microbiota. Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, and Gilad Y. PLoS One. 2015;10(11):e0140301
This article was highlighted as an Editor’s pick for both the PLoS Microbiology and PLoS Experimental Biology special collections:
http://collections.plos.org/microbiology-picks &
http://collections.plos.org/experimental-biology
Also check out our GWAS of the nasal microbiome in the Hutterites, led by Dr. Catherine Igartua:
Host genetic variation in the mucosal immunity pathways influences the upper airway microbiome Igartua C, Davenport ER, Gilad Y, Nicolae DL, Pinto J*, and Ober C*. Microbiome. 2017;5(16)
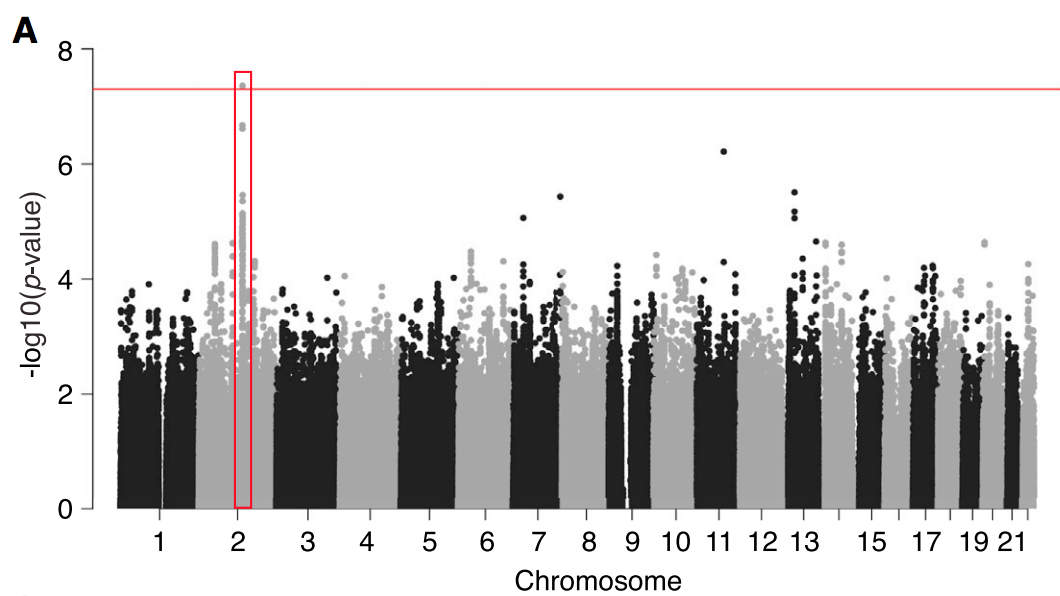
«GWAS of gut microbiota in TwinsUK»
GWAS in the TwinsUK
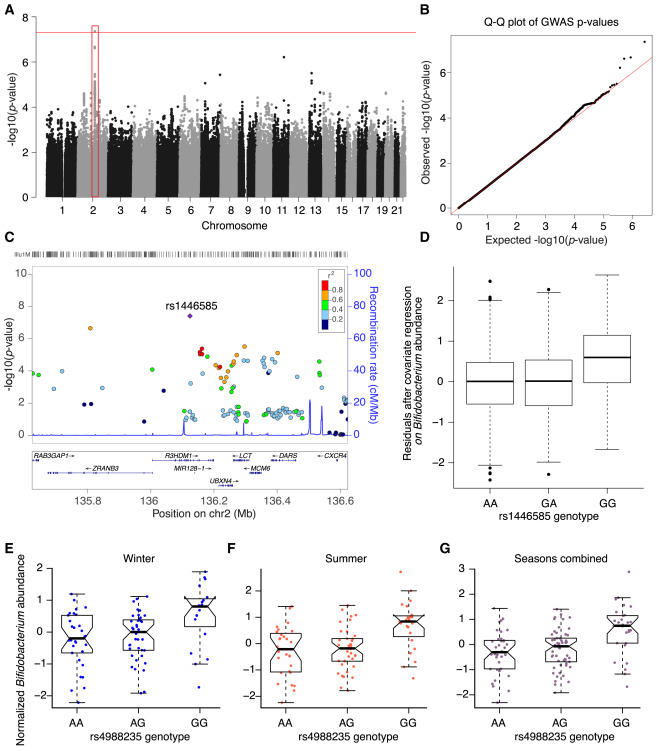
As a postdoc, I continued examining the role of host genetics using the TwinsUK cohort.
This large, well-characterized cohort gives us much more power to detect associations between variation in the human genome and abundance of bacteria in the gut. We now have replication across multiple studies that show lactase persistence alleles are associated with lower levels of Bifidobacteria in the gut.
The study was led by Julia Goodrich, while I contributed several analyses to study, including the replication of the LCT-Bifidobacterium association in the Hutterites, PrediXcan analyses, and microbiomeGWAS analyses.
To read more, check out:
Genetic determinants of the gut microbiome in UK twins Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, and Ley RE. Cell Host & Microbe. 2016;19(5), 731-743
Complex genotype associations with gut microbiota in TwinsUK
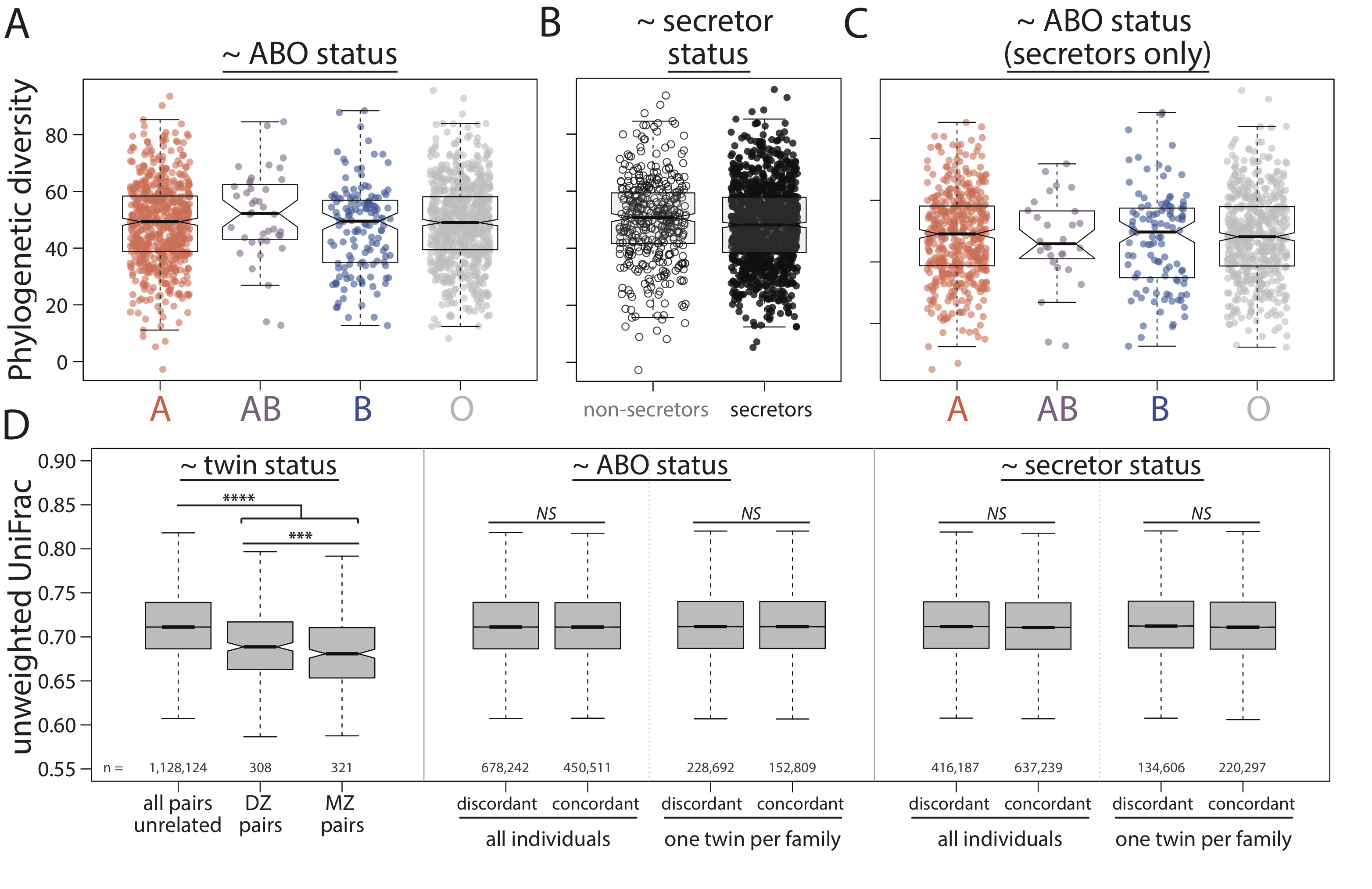
In addition to genome-wide studies in the TwinsUK, I also examined two loci in depth that were previously implicated as playing roles in determining gut microbiome composition.
Cell surface antigens potentially act as either energy sources or tethers for bacteria to adhere to the gut.
Therefore, host variation controlling antigen structure or expression could be factors influencing gut microbiome composition.
One such antigen is encoded by the ABO locus - known commonly as your blood type.
This antigen not only exists on the surface of red blood cells, but also along mucosal surfaces in individuals with an intact FUT2 gene (secreting individuals).
ABO antigen and secretor status are risk factors for a number of diseases, including some that also show association with gut microbiome composition.
With that in mind, I set out to identify links between the host genome (in ABO and FUT2), the composition of the gut microbiome, and disease state.
However, that plan was cut short as ABO and secretor status were NOT associated with overall microbiome composition, diversity, or the abundances of individual bacteria in the gut, despite the barrage of different tests thrown at the data.
These [lack of significant] results highlight the importance of performing replication studies using large, well-powered cohorts.
To read more, check out:
ABO antigen and secretor statuses are not associated with gut microbiota composition in 1,500 twins. Davenport ER, Goodrich JK, Bell JT, Spector TD, Ley RE, and Clark AG. BMC Genomics. 2016;17:941
«Reviews and commentaries»
In addition to examining host genome-microbiome interactions in the two study populations highlighted above, check out some reviews and commentaries I’ve written on the subject:
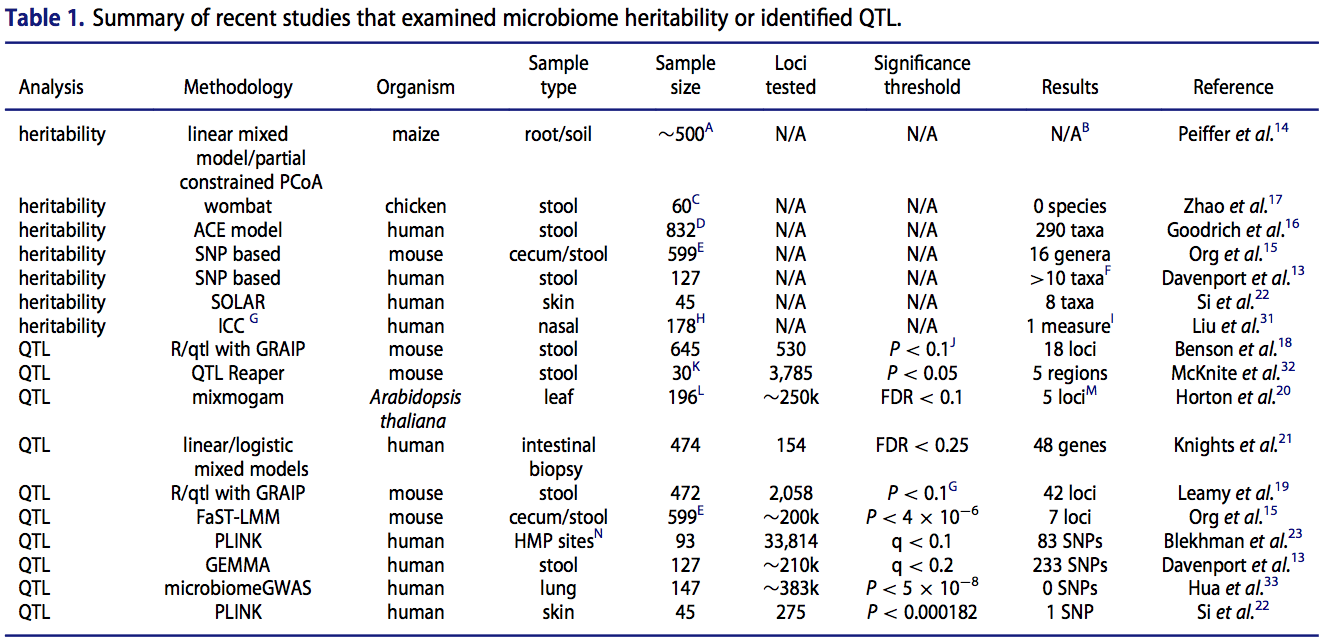 Elucidating the role of the host genome in shaping microbiome composition.
Elucidating the role of the host genome in shaping microbiome composition.
Davenport ER. Gut Microbes. 2016;7(2), 178-184
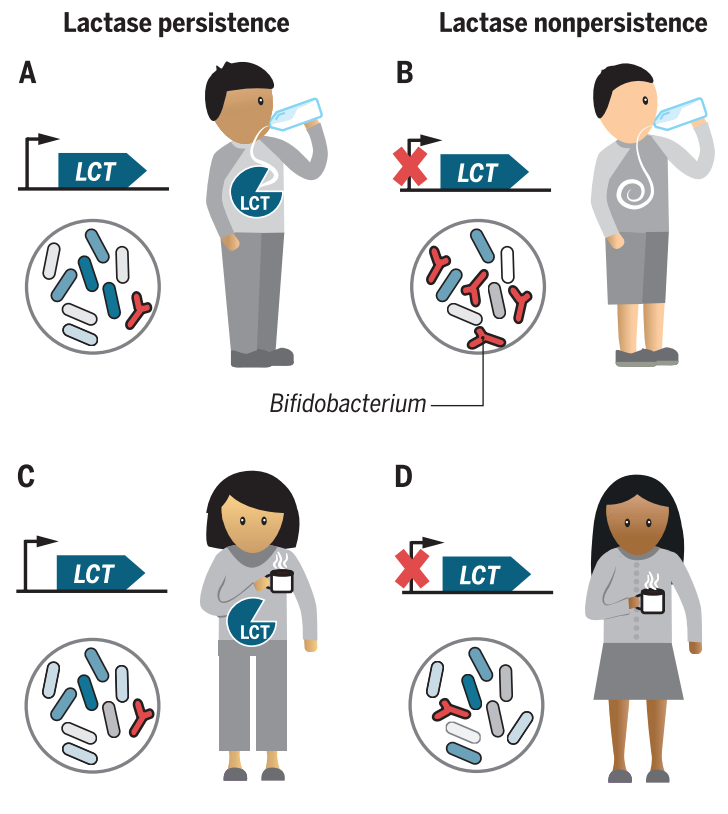 Cross-species comparisons of host genetic associations with the microbiome
Cross-species comparisons of host genetic associations with the microbiome
Goodrich JK*, Davenport ER*, Waters JL*, Clark AG, and Ley RE. Science. 2016;352(6285), 532-535
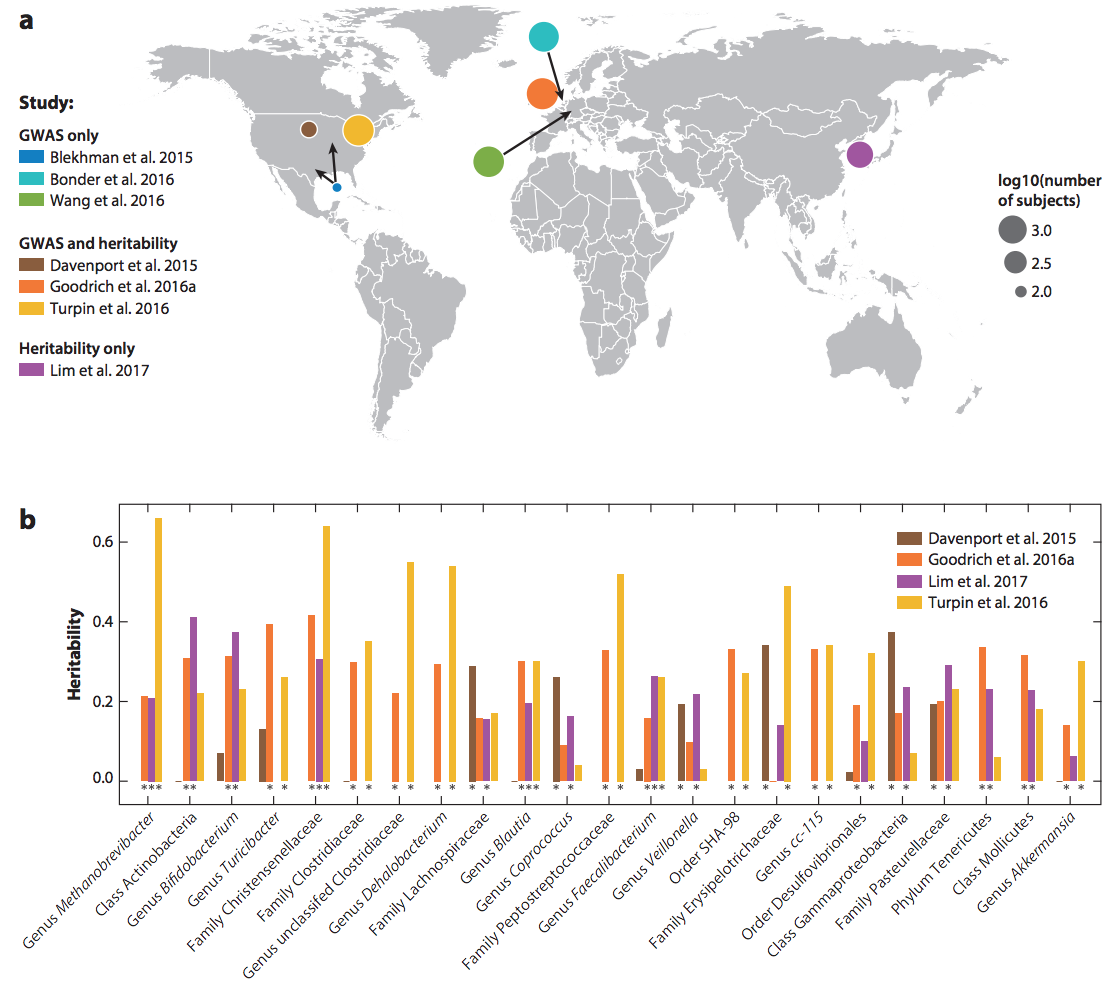 The relationship between the human genome and microbiome comes into view
The relationship between the human genome and microbiome comes into view
Goodrich JK, Davenport ER, Clark AG, and Ley RE. Annual Reviews of Genetics. 2017;51, 413-433
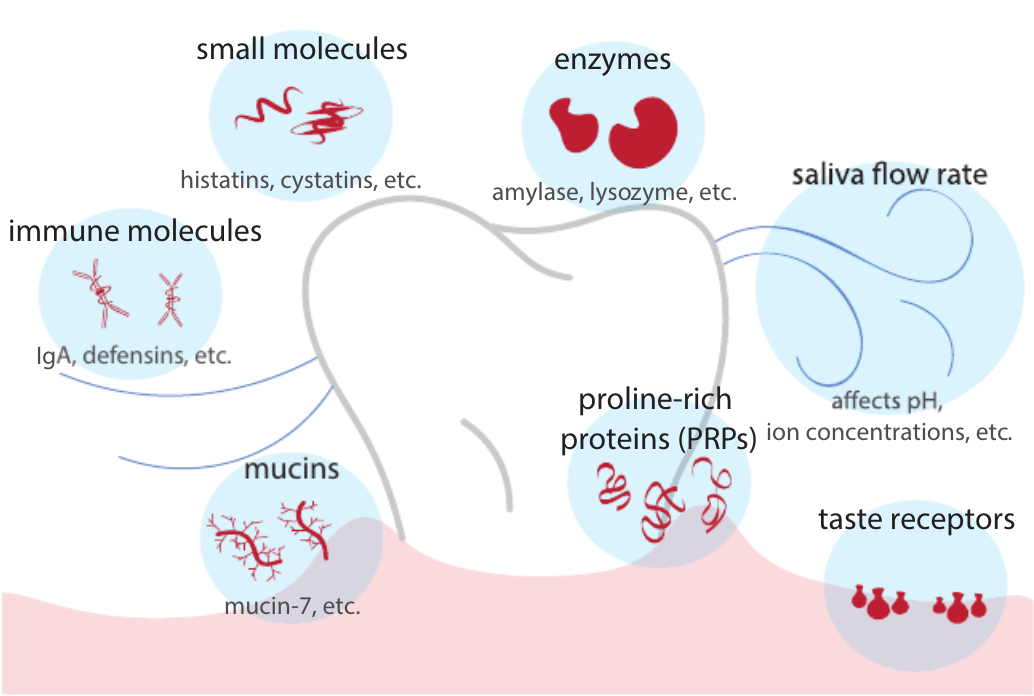
 Tooth be told, genetics influences oral microbiome
Tooth be told, genetics influences oral microbiome
Davenport ER. Cell Host & Microbe. 2017;22(3), 269-278