Microbiome method development
Microbiome research is a relatively new and rapidly expanding field. New wet lab and statistical approaches are needed to answer fundamental questions about host + microbe interactions. Here’s how we are contributing.

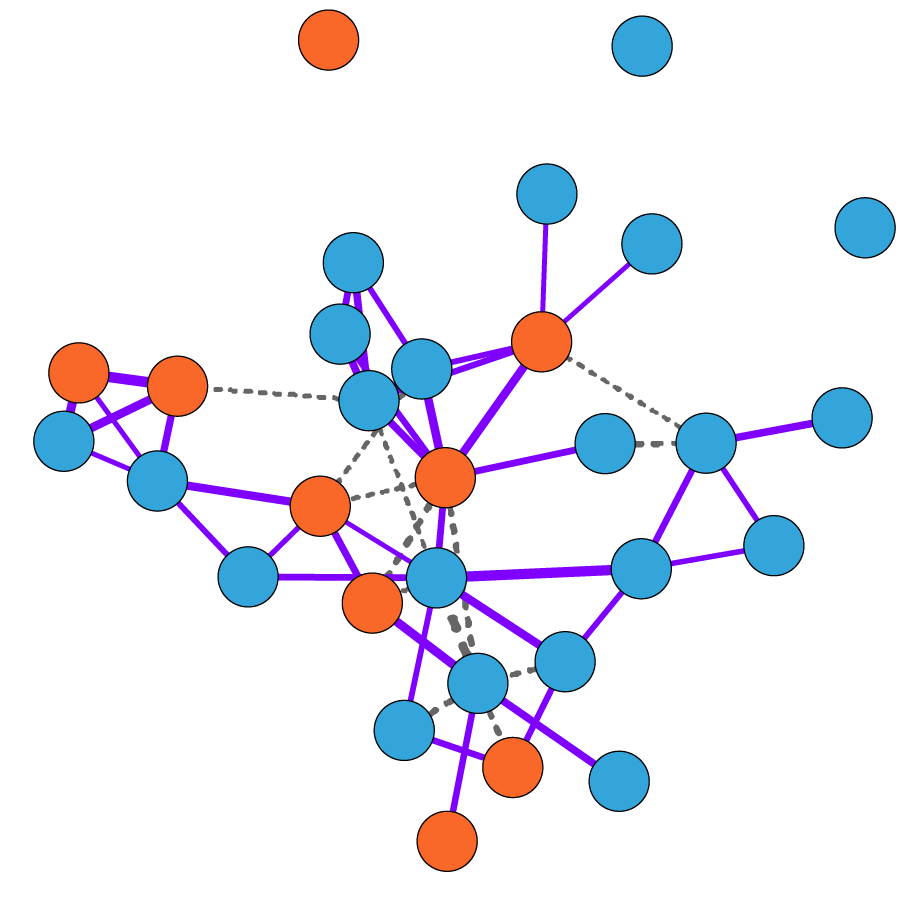
Co-occurrence network modeling reveals disease-specific configurations of microbiome community structure
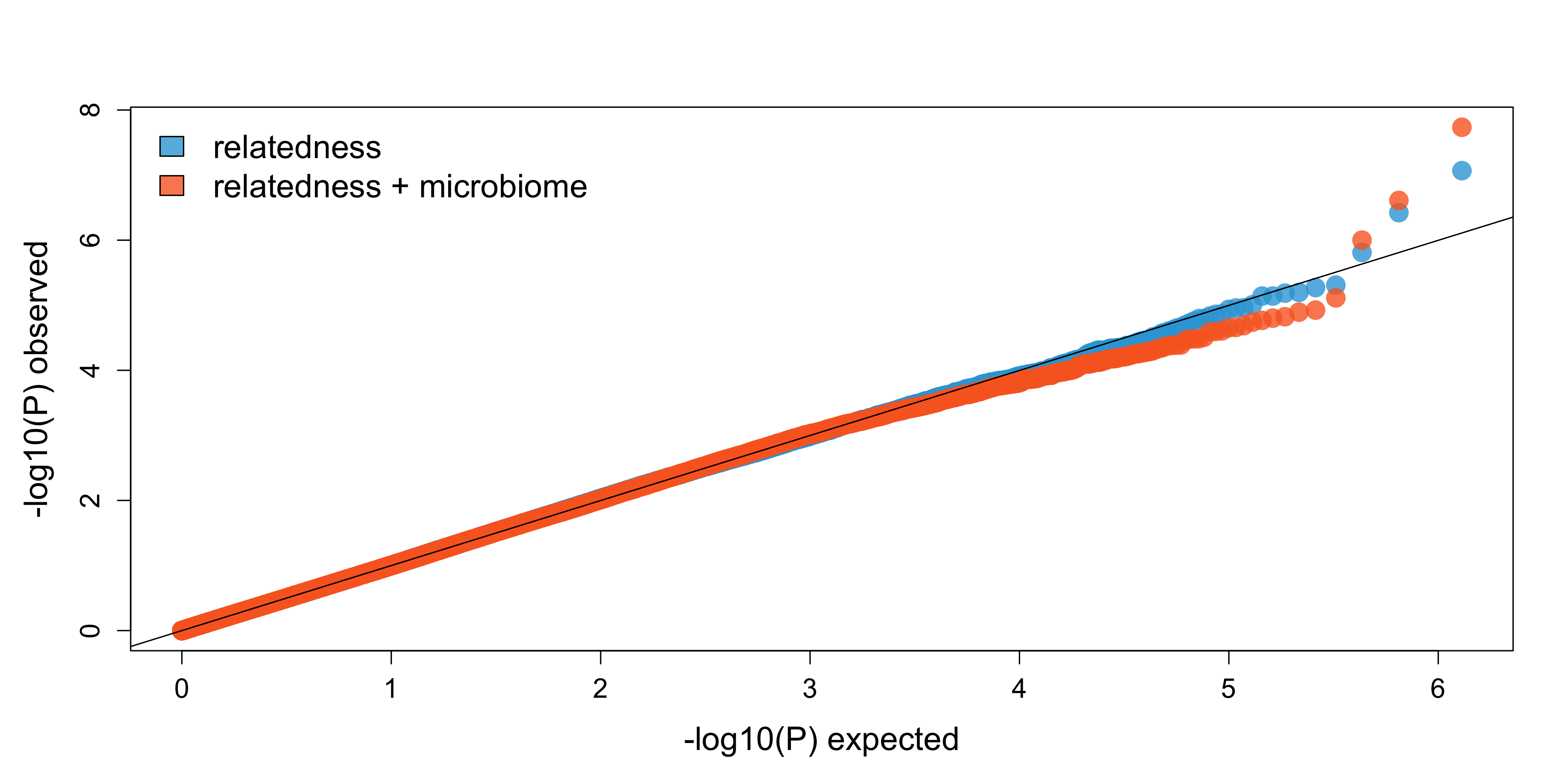
Simultaneously modeling host genetics and microbiome composition reveals the heritability and the proportion of variance explained due to the microbiome of immune-related traits
Co-occurrence network modeling reveals disease-specific configurations of microbiome community structure

The human gut microbiome is associated with a large number of complex traits and diseases. Although certain bacterial taxa have been identified that correlate with disease, it is commonly thought that alterations in microbial community dynamics are indicative of a disease state. To investigate this, we are examining how microbial community structure shifts between healthy and disease states for ~180 immune-related diseases and quantitative traits in a cohort of 2,500 twins from the United Kingdom using systems biology techniques. Using co-occurrence networks, we are investigating both whether network-level properties differ between i) taxa associated with disease (centrality, connectedness, etc.) and ii) whether overall topology differs between networks built with affected or unaffected individuals (modularity, transitivity, etc.). Our goal is to identify consistent changes in the microbiome community structure in health versus disease, across either all or a subset of phenotypes.
This work was presented at the following conferences:
Biology of Genomes (BoG) - 2017
Society of Molecular Biology and Genetics (SMBE) - 2017
American Society of Human Genetics (ASHG) - 2017
Simultaneously modeling host genetics and microbiome composition reveals the heritability and the proportion of variance explained due to the microbiome of immune-related traits

Both host genetics and environmental factors determine human immune-related phenotypes and disease, including asthma, allergies, and rheumatic disorders. Although many known environmental exposures contribute to immune disease risk and severity, the extent to which the human microbiome is responsible for variation in these phenotypes remains largely unknown. Additionally, whether gene-by-environment interactions with the microbiome influence these phenotypes remains uncharacterized, as well as which specific genetic variants and microbes play roles in this process.
To address these gaps, we are examining both the proportion of variation explained by host genetics (PVE-G, or heritability) and gut microbiome composition (PVE-M) in a unified framework for ~30 immune-related phenotypes using the TwinsUK cohort. Additionally, we are exploring whether explicitly including gut microbiome composition in genome-wide association study (GWAS) models improves power to detect genetic variants associated with immune phenotypes.
This work was presented at the following conferences:
American Society of Human Genetics (ASHG) - 2018
Probabilistic Modeling in Genomics (ProbGen) - 2018